Published today in PLOS Pathogens, a massive collaboration lead by graduate student Aaron Embry joint-appointed to Alto and Gammon labs identifies a new proteins that participate in the regulation of viral replication in host cells. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1012010
Category Archives: Publications
Crystal Structures of Ceg10 – A labor of Love
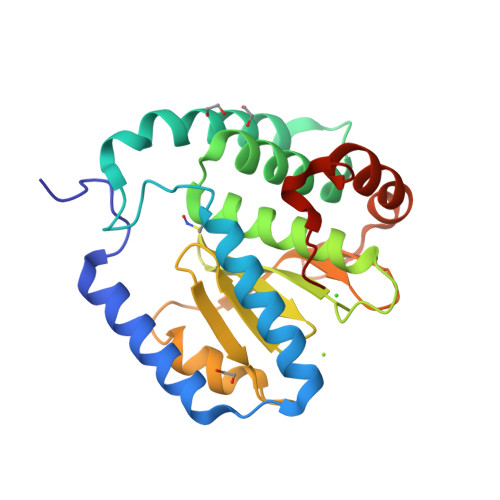
Today, the crystal structure of Ceg10, an effector protein from Legionella was deposited and released at the RSCB Protein Data Bank (PDB). While a lot remains to be determined about its function and targets during an infection, we are excited to finally release some of what we have learned about this protein in the last 8 years! The Ceg10 crystal structures are part of a collaborative project between Dr. Heisler’s lab and the Gammon, Alto, and Taglialucci lab’s at UT Southwestern Medical School.
More about what Ceg10 does during a viral infection can be found in a recent preprint: Exploiting Bacterial Effector Proteins to Uncover Evolutionarily Conserved Antiviral Host Machinery
The crystal structures can be found at the RSCD PDB: 9B8D and 9B8E
New Paper out in Nature Microbiology
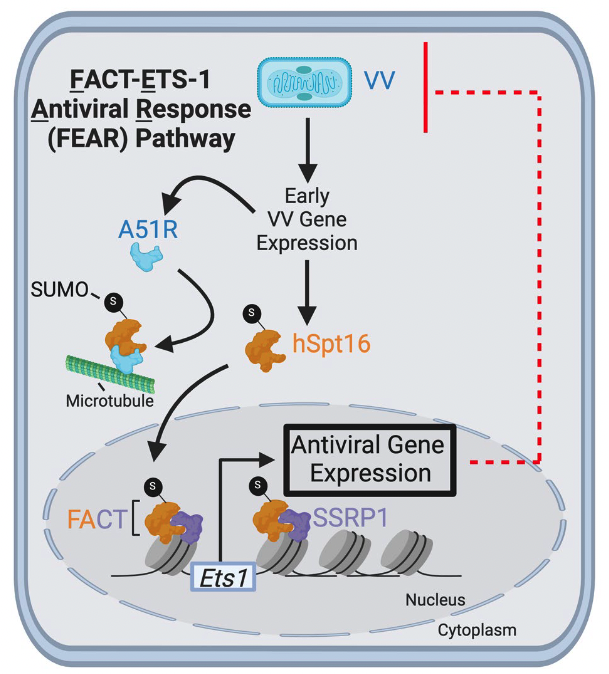
Published today in Nature Microbiology, a massive collaboration lead by “fear”less graduate student Emily Rex in the Gammon lab identifies a new pathway for restricting viral pathogens and how some viruses attempt to evade this pathway. More can be found at: https://www.nature.com/articles/s41564-024-01646-5.epdf?sharing_token=scTW3uKvkhn77dxbSmqu59RgN0jAjWel9jnR3ZoTv0PtJOB0uazhwtp9ry6BHEUNjpgTszmdaCSu_Nf9eB9lAyknydEiu__2sDUwM0iG8EUbIJGY3RbQovhdc6-guMR3hCQJ2aNLgP1odVuOxusrG-J3kvf2297z1uGSL2XXcQA%3D
Latest Pre-Print From Dr. Heisler’s Postdoc is online
Released today on bioRxiv, the Alto and Gammon labs identify previous unknown host factors that restrict viral replication during cellular infections. Led by graduate student Aaron Embry, this study used a library of bacterial effector protein genes, which Dr. Heisler generated, to screen for proteins that suppressed host immune factors and allowed an otherwise restricted virus to replicate in cells. One effector protein, called Ceg10, that rescued all of the viruses tested, was shown by Dr. Heisler to have a Cys-His-Asp catalytic active site and, using limited proteolysis and X-ray crystallography, a high resolution structure of the core domain of the protein was solved. Additional work remains to be done to determine exactly what Ceg10 does and what its host target(s) is, but a lot of promising work is underway to figure this out!
Abstract:
Arboviruses are a diverse group of insect-transmitted pathogens that pose global public health challenges. Identifying evolutionarily conserved host factors that combat arbovirus replication in disparate eukaryotic hosts is important as they may tip the balance between productive and abortive viral replication, and thus determine virus host range. Here, we exploit naturally abortive arbovirus infections that we identified in lepidopteran cells and use bacterial effector proteins to uncover host factors restricting arbovirus replication. Bacterial effectors are proteins secreted by pathogenic bacteria into eukaryotic hosts cells that can inhibit antimicrobial defenses. Since bacteria and viruses can encounter common host defenses, we hypothesized that some bacterial effectors may inhibit host factors that restrict arbovirus replication in lepidopteran cells. Thus, we used bacterial effectors as molecular tools to identify host factors that restrict four distinct arboviruses in lepidopteran cells. By screening 210 effectors encoded by seven different bacterial pathogens, we identify six effectors that individually rescue the replication of all four arboviruses. We show that these effectors encode diverse enzymatic activities that are required to break arbovirus restriction. We further characterize Shigella flexneri-encoded IpaH4 as an E3 ubiquitin ligase that directly ubiquitinates two evolutionarily conserved proteins, SHOC2 and PSMC1, promoting their degradation in insect and human cells. We show that depletion of either SHOC2 or PSMC1 in insect or human cells promotes arbovirus replication, indicating that these are ancient virus restriction factors conserved across invertebrate and vertebrate hosts. Collectively, our study reveals a novel pathogen-guided approach to identify conserved antimicrobial machinery, new effector functions, and conserved roles for SHOC2 and PSMC1 in virus restriction.
To read more, visit https://www.biorxiv.org/content/10.1101/2024.01.29.577891v1